Hi everyone, I am Eleen – (soon-to-be) second-year master student at Columbia University Ecology, Evolution, and Environmental Biology department. I study ecosystem ecology, broadly interested in nutrient cycling. My current work is focusing on Nitrogen (N) cycling in temperate forest ecosystems.
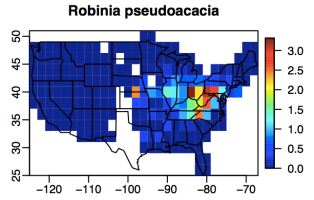
One of the major supply of N in terrestrial forest ecosystem is N fixation by free-living or symbiotic microbes that live in the root nodules of N-fixing plants. My research is primarily focusing on symbiotic N fixation (SNF): SNF is a process of converting atmospheric N2 into plant-available N by microbes. It is crucial in global n cycle, especially important in temperate forests, where N commonly limits primary production. SNF is also essential in conservation because of its capability of jump-starting recovery in abandoned and nutrient-poor systems to facilitate nutrient restoration and succession. In coterminous U.S., the estimated N input combining both N deposition and other types of N fixation besides symbiotic N fixation is <20kg N/ha year (Holland and Braswell 2004, Reed et al 2011). Based on a rough estimate, N fixers have the potential to be the greatest N input source on a continental scale (Sprent and Parsons 2000). Based on the recent Forest Inventory Analysis (FIA) data analysis (Liao and Menge Unpublished), there are in total 12831 N fixing trees recorded since 1980s. Genus Robinia (Figure 1) is the most common of six native N-fixing genera across U.S.: on a regional scale, this genus is particularly important in northeast region (Figure 2) where it comprises 95.6% of N fixer biomass. Despite the recognized importance, we currently do not have an accurate understanding of SNF rates of genus Robinia in this region or how they vary with tree age. My summer plan is to conduct a systematic analysis of Robinia SNF rates across tree ages in a northeast temperate forest.
(Figure1: Robinia at Central Park)
(Figure 2: Distribution of Robinia across U.S. Color shows percent basal area. (Menge et al 2010))
There are different ways of measuring SNF rates- each has advantages and limitations. The most commonly used methods include 15N enrichment experiment, acetylene reduction assay, and d15N natural abundance method. 15N enrichment experiment allows ecosystem-level analysis but is high-costly. Acetylene reduction assay because of its invasive nature by excising nodules off might not capture the actual SNF rates under natural settings. In my summer research, a combined analysis of 15N isotope natural abundance and dendrological dating record will be used to approach SNF rate of Robinia across age groups. I will sample 15 pairs of adult Robinia trees and neighboring non-N-fixing trees at Black Rock Forest (BRF), assuming they access the same soil nutrient pool. By coring the trees, dividing tree cores into different age groups, and comparing the isotope signals of Robinia and reference non-N-fixing tree I will be able to understand the amount of N fixed and the rate of SNF of genus Robinia in this forest and thus provide an estimate of SNF rates in northeast region.
Before going down to the field and start coring trees, I will spend/have been spending my time compiling literature that have reported SNF rates of Robinia and other N-fixing genera across coterminous U.S. In addition to literature review, I also will continue figuring out the best way to extract isotope signals from tree cores. Ultimately the data with literature estimates of SNF rates and experimental measurements of SNF rates will be combined with FIA database to estimate SNF at a continental scale, providing the first ever map of nationwide SNF estimates.
Reference:
Holland, E. A., B. H. Braswell, J. Sulzman, and J.-F. Lamarque. 2004. Nitrogen Depositio to the United States and Western Europe. Data set. Available on-line [http://www.daac.ornl.gov] from Oak Ridge National Laboratory Distributed Active Archive Center, Oak Ridge, Tennessee, U.S.A.n on
Menge, D.N.L., DeNoyer J.L., and Lichstein J.W. 2010. Phylogenetic constraints do not explain the rarity of nitrogen-fixing trees in late-successional temperate forests. PLoS ONE 5(8): e12056.
Reed, S. C., C. C. Cleveland, and A. R. Townsend. 2011. Functional Ecology of Free-Living Nitrogen Fixation: A Contemporary Perspective. Annual Review of Ecology, Evolution, and Systematics 42:489–512.
Sprent, J., and R. Parsons. 2000. Nitrogen fixation in legume and non-legume trees. Field Crops Research 65:183–196.