The latest installment of Breaking Bio interviews has arrived! Check out this mashup of our very own mammal-loving Masters’ students, Kaggie Orrick, Jess Stitt, Erin Dimech, and Allison Roth:
The latest installment of Breaking Bio interviews has arrived! Check out this mashup of our very own mammal-loving Masters’ students, Kaggie Orrick, Jess Stitt, Erin Dimech, and Allison Roth:
Read about MA second-year Erin Eastwood’s genetic work with Fijian fishes!
Over the last two months I have transformed from a perpetually damp, wetsuit-clad, village-hopping field scientist, into a (relatively) well-dressed, city-living, pipette-wielding lab scientist. Where I used to spend most of my time chasing fish around with my spear and catch bag, my life now largely consists of moving tiny amounts of liquid from one tube to another. And surprisingly enough….it’s pretty darn awesome.
 Step into my office.
Step into my office.
Lab work is enjoyable to me for two main reasons – first, I am constantly in awe of the fact that we humans have even FIGURED OUT how to extract, amplify, and sequence DNA from living tissue. The decades of brainpower, creativity, and technological innovation required to make those little A’s, G’s, C’s, and T’s come up so nicely on our computer screens is boggling, and keeping that in mind brings a certain kind of appreciation to even the most tedious tasks at the bench. That’s not to say it’s always totally easy and…
View original post 317 more words
Now that we’ve reached mid-August, most of us MA students are returning from summer fieldwork and re-settling into life at Columbia. (Or, if you were hanging around NY this summer like me, you’re getting excited to have the department bustling again.) We’re now looking ahead to the fall: application season and thesis-writing extravaganza. But, the summer’s been pretty productive.
I’ve almost finished sequencing the glucocorticoid receptor gene in 24 African starling species. (I’ve had to re-design primers for an exon that sequenced poorly, despite clear bands on the gel.) Thanks to the photography help of the lovely Laura Booth (an undergraduate in the Rubenstein lab who has blogged about her work this summer here), I’ll show my workflow from DNA to sequence data.
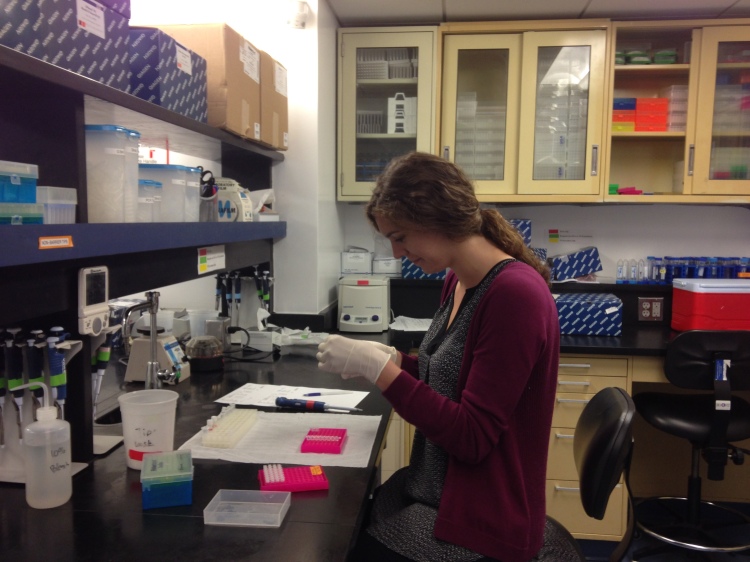
Checking DNA aliquots.
Step 1: Copy DNA (PCR)
Using the primers I designed last spring, I ran many, many PCRs (Polymerase Chain Reactions) to amplify tiny DNA fragments that range from 100-1200 base pairs in length. To prepare a PCR, I first pipet the template DNA, and then add a “mastermix,” which is a combination of molecular water, Tris buffer, magnesium chloride, primers, Taq polymerase and dNTPs (deoxyribonucleotides, or the actual “ATCGs” that the polymerase needs to copy the template DNA). Then, I load the PCR onto a thermocycler and wait for 1.5-2 hours.

Putting the PCR on the thermocycler.
Step 2: See If Step 1 Worked (e.g. Gel Electrophoresis)
Next, I run that PCR product out on an agarose gel to see if I actually copied the DNA. The gel is an agarose sugar suspended in 1xTAE buffer, which creates a grid through which the DNA “runs.” On each gel, I add a ladder that shows bands of specific sizes; by comparing the position of each band to the ladder, I can check that my product is the correct size (e.g. 585bp for Exon 8 of NR3C1). I also check that each band is bright and clear, to be sure that there is enough PCR product and that all of that product is the same size. Running gels is pretty satisfying: once you get the hang of loading the dye-PCR mixture into the wells without touching the gel bed or introducing bubbles, it becomes pretty robotic.
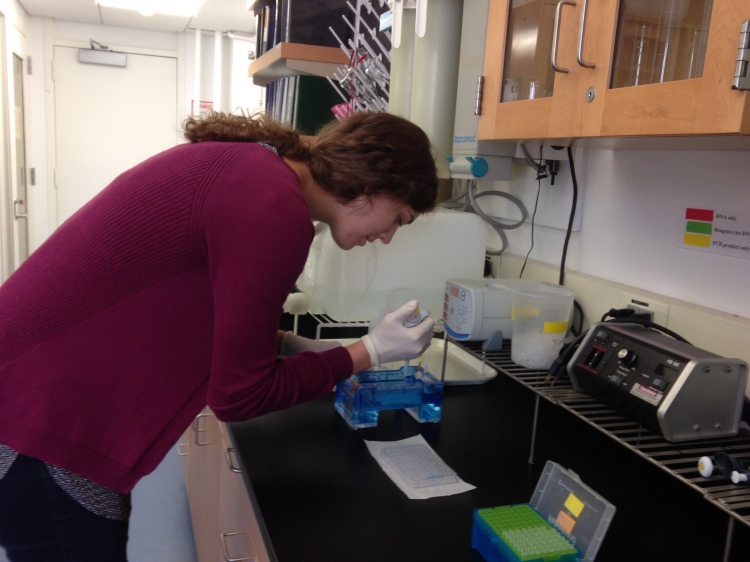
Loading a gel.
Step 3.5: Do Step 1 Again, and Hope Hope Hope It Works
Given that I only had about 40 individuals and 8 exons to sequence, I hoped to finish this part of my thesis by early July, but science rarely works out like that. I’ve run three to four rounds of re-dos (which is normal), in order to finish 7 of the 8 exons. I’m still working on Individual 5-3 in Exon 8, a Lamprotornis regius that hasn’t been cooperating, but that guy should be sequenced tomorrow. I’ve also had to re-design primers for Exon 3, and I’m prepping the sequencing reaction for the finicky 5-3 and another round of Exon 3 in a few minutes.
Step 3: Sanger Sequencing
I use a three-step process for sequencing: two clean-up steps on either end of a cycle sequencing reaction. First, I use two enzymes (Exo and SAP) to clean the PCR product to dephosphorylate and remove free dNTPs and single-stranded DNA. Then, I mix the cleaned PCR product with more primers, sequencing buffer, molecular water and Big Dye (a stress-inducing expensive dye) and load this reaction onto the thermocycler for 3 hours. Finally, I spin that cycle sequencing product down through a hydrated matrix of Sephadex powder to remove any unused dye-labeled nucleotides. Then, I’m ready to take it down to the ABI 3730 sequencer at the museum and wait for my data.
Since the Big Dye is light-sensitive, I do this reaction in the dark (because I’m overly cautious and don’t want to risk losing precious DNA due to a failed sequencing reaction). So, I’ll leave it to you to imagine what the steps listed above might look like (complete with my requisite lab-hunching that I worked pretty hard to correct in the pictures you’ve seen – don’t worry, I have plenty of pictures of what I actually look like in lab.) I’m now off to hide out in the dark again, and hopefully, if the Science gods are kind, finish the very last round of sequencing!
See some of Erin E’s photos from Fiji fieldwork!
Some glorious photos from Kaggie, conducting her fieldwork in South Africa!
This past month I have been working at Black Rock Forest, a research and educational forest located in Cornwall, NY. I am interested in the changes in mesofauna, soil-dwelling animals that are about a few millimeters in size, to forest disturbances. If you want to learn more about soil biodiversity and why my project is important, you can watch my video featured in Breaking Bio Blitz in the previous post. This post will focus on my fieldwork and the methods I have been using to collect all the animals living on the soil.
The forest disturbance part of my project was inspired by an on-going long-term experiment at Black Rock Forest. Named the “Future of Oak Forests Study,” this project was initiated in 2008, and includes different tree girdling experiments, with an emphasis on the girdling of red oak trees to simulate an increasingly common scenario where sudden oak death takes over a forest. A smaller portion of this experiment includes herbivore exclosures, where parts of the forest are fenced off in order to prevent feeding by large and medium herbivores which, at Black Rock, are deer. You can read about the experiment here.
Photo: A girdled red oak tree, Black Rock Forest, October 2013
Working in these experimental plots is a unique experience. In certain areas, you are surrounded by girdled trees, which are trees that have been sawed around the trunk at breast height so the trees are still standing but are functionally dead. In herbivore exclosures, the vegetation, light intensity and overall appearance of the forest is very different.
Many studies in this experiment area have obviously focused on the aboveground response of vegetation to the removal and death of oaks and the removal of major herbivores. I am interested in what was going on in the soil and leaf litter. In order to collect mesofauna, which are so small that you can only really see their general form with the naked eye, I had to collect leaf litter at the surface of the forest floor and soils at different depths. I used a soil corer, which is made up to two, very heavy parts: a slam hammer, to drive down into the soil and a metal cylinder, to collect the soil. By placing the metal cylinder upright on top of the soil, after the leaf litter has been removed, I lift the hammer and drive it down into the ground about 10 centimeters, which is sometimes a lot more difficult than one would expect. Once the cylinder was full of soil (and not rocks, which I found to be very problematic in certain areas of the forest), I had to gently extract it from the ground and place the soil into (properly labeled) bags. I took many, many cores in different part of the experimental area for both mesofauna collection as well as soil physical and chemical properties analysis.
Photos: My Berlese funnel set-up, Black Rock Forest, July 2014
Once I collected all my soils and leaf litter, the mesofauna had to be collected from the different mediums. The outstanding and universally applied method of doing this type of arthropod extraction is a Berlese funnel. The principle of a Berlese funnel is to use both heat and light to drive the animals out of the medium and into vials of ethanol to be collected and sorted later. The leaf litter and soils were placed into large funnels resting below a light bulb, the source of heat and light, with the end of the funnel falling into a vial of ethanol. By letting the soils and leaf litter dry out for three days, I expected to get most of the arthropods and other animals to migrate away from the light bulb and fall into the ethanol. As of right now, all my soils and leaf litter have been “extracted” and I have a bunch of ethanol vials filled with mesofauna and other animals and arthropods to be counted and sorted. I’m hoping to be able to identify the major mesofauna groups (as well as a few subgroups) and determine whether or not the forest disturbances are having a significant effect on these communities.
Though I’m sad that my fieldwork and funnel days are over, I am looking forward to seeing all the litter critters that have been living in the leaf litter and soil under the microscope as I start the next part of my summer research.
-Natalie Bray
Another Breaking Bio Blitz video featuring Natalie Bray, talking about the importance of soils and soil biodiversity.
Bula everyone! Erin here from Fiji, wanted to post this little GoPro video I put together from one of our field trips over the last week. My advisor, Josh Drew, and I have been teaching a marine conservation course at USP with students from Fiji, Vanuatu, Papua New Guinea, New Zealand, and Germany, and on Wednesday we went for a snorkel to look for examples of symbiosis on the reef! Check it out to see what we found…so much fun!
Greetings to my fellow Masters students working hard on summer research and also to all you kind readers interested in our adventures! I am writing from the town of Mundemba in Southwest Cameroon, where I have been based for just a little over a month now for my research on human-wildlife conflict. I had hoped to write earlier, but in just the first week my computer was tragically killed during one of the frequent power surges. Electricity is patchy and internet nearly non-existent. But, alas, I am writing now – here and well and better adjusted to the trials and tribulations of research in Cameroon!
For those of you not familiar, my research is centered on understanding human-wildlife conflict in the form of crop raiding within the context of existing and increasing oil palm development in Cameroon. Oil palm production has gained international attention in recent years due to the extensive deforestation and human displacement caused by plantation expansion in Indonesia. Now, many of the same Southeast Asian companies operating in Indonesia, in addition to new oil palm companies from the U.S. and Brazil, are looking to the forests of West and Central Africa as a new frontier for expansion. There are many reasons why large-scale oil palm production will differ between Indonesia and Africa, way too many to address here. Most significantly, the oil palm tree (Elais guineas) is native to Africa, so palm oil and its by-products hold significant cultural relevance bolstered by a long legacy of smallholder production. I chose to conduct my research in Cameroon, a country where many industrial oil palm concessions are in the works, because I believe the potential impact of these companies on human-wildlife conflict surrounding forested areas needs to be addressed in the African context.
My work here in Cameroon involves visiting four different villages at varying distances from two protected areas – Korup National Park and the Rumpi Hills Forest Reserve – and two oil palm production sites – one a long-standing government-owned oil palm plantation called PAMOL and the second a recent concession acquired by SGSOC (Sithe Global Sustainable Oils Cameroon). Within each village, I spend two weeks interviewing households chosen through systematic random sampling of homes. Each interview involves a set of around 40 questions regarding basic household information, diet (particularly bushmeat consumption), farming, and perspectives on land use change. The section on farming is the most substantial part of the interview. I ask about the different animals that visit the participant’s farm, how frequently, and the extent of damage for each animal. I then schedule a time to visit the farm and take a GPS point so that I can later uncover spatial predictors of crop raiding.
So far, I have completed a pilot study in the village of Meka Ngolo, two weeks in the village of Mundemba II, and am now wrapping up my first week in the village of Ikondo-kondo. Each village has proved to be an enriching experience for my perspective of conservation science in practice, as well as my personal development. I have been received with welcoming arms and in many cases, enthusiasm, because my research indirectly provides an outlet for them to express the daily grievances they face with the animals damaging their farms. As the majority of people in these villages identify as farmers, the damage caused by animals on farms often has significant consequences for household income and subsistence. Thus, it is not surprising that the direct, negative impact of animal raiding behavior on livelihoods often results in lethal action.
As I carry out my research, there have been some really interesting developments. I am not able to describe all of them just yet (without some preliminary analysis!), but one revelation concerns how deeply entrenched human-wildlife conflict is with broader elements of land use and tenure in this region. Both Meka Ngolo and Mundemba II share close boundaries with the Rumpi Hills Forest Reserve and the PAMOL plantation – two vastly different land use types with their own set of restrictions to farming. The village I am currently working in, Ikondo-kondo, was resettled out of Korup National Park fourteen years ago to a new area where crop-raiding by elephants is on the rise. And so, as goes the scientific process, I am working on adjusting my framework to incorporate the critical, changing dynamics of land use.
All in all, I am finding my time here to be incredibly rewarding. The work has certainly not been easy. Interviews often require tricky translation into Cameroonian Pidgin and local dialects. Visiting farms often involves trekking up hills that feel as if they are on a 90 degree incline, crossing wide streams and failing to keep my wellies dry, and trotting through thick farm bush that is known to hold Rhinoceros vipers and pythons. However, at the end of each day I remember that I am very privileged to be here – learning from the rich, bright culture of Cameroon while trying to make a dent in conservation practice through my work!
-Cynthia Malone
Out of the classroom and into the field! Summer is all about learning in unexpected settings and in new ways.
Hi! I’m Amy– a grad student in Dr. Josh Drew’s research group here in Columbia’s E3B Department. My research focuses on the diversity of coral reef fish in the Indo-Pacific Ocean. I’m interested in how these fishes have evolved, and how they have migrated over generations to populate the reefs they now call home.
Lately I’ve also been thinking a lot about science communication– how do we as a community share our research with a broader audience to increase enthusiasm (and funding) for science? And moreover, for the 9 out of 10 graduates who won’t find a tenure track job in academia, how do we parlay our salmagundi of skills into employment?
The majority of the year, I’m right here in New York, tackling the beast known as thesis research, quietly repressing thoughts of my next career move. But summer is a special time. It frees up a few weeks to mix up the normal routine, get out of town, and pursue new ideas in far-flung locales.
So, spicing up my summer work regimen, I headed to exotic Cambridge Massachusetts last week for ComSciCon 2014 –a conference all about communicating science! Over four full days, 50 graduate students met for a series of workshops on the effective communication of science to non-scientist audiences.
This conference is centered on a series of interactive panels, where policy makers, journalists, and academic research scientists discuss the state of science communication today. Central questions included: How is science portrayed differently to lawmakers on Capitol Hill, or to the general public, than it is to graduate students or research scientists? How do scientists perceive STEM, and are we effectively communicating our vision to broader audiences?
While we talked about communicating science, we also talked about working in science. And after each panel, I schmoozed; I dug; I generally begged for advice. Here is what I found– my top 5 insights from ComSciCon 2014:
A graduate degree is not a silver bullet
Duh. These days credentials aren’t enough– your experience, skill set, and network matter tremendously too (more on this in a moment). But for students in the STEM fields, that realization comes as a particularly painful slap in the face. We used to have it good, man– a grad degree once translated to relative job security– but alas, no more. Now, at best, 1 in 10 PhDs will find a tenured professorship, and unfortunately, there isn’t a textbook on the way forward for everyone else.

Outside of the ivory tower, career paths are numerous and decentralized. That well trodden pathway of PhD à post doc (or three) à assistant professor à tenure forever (!?) is actually quite unusual in it’s linear clarity.
A few universities are responding by creating graduate career counseling offices– previously unheard of in academia (Check out UC Berkeley’s Beyond Academia for one example). Resources for non-academic job placement are certainly growing as academic positions dwindle, but clearly outlined stepping stones to alternative careers are still a long way off.
Networking is (almost) everything
So, there’s no silver bullet. Bummer.
But, after schmoozing ComSciCon panelists (ostensibly very successful people) around the hors d’oeuvres for four days straight, I did come away with one majorly helpful realization: the common denominator between all these employed people is a kickass network.
Panelists overwhelmingly cited networking and luck as the major factors that helped them get their start. And that makes sense doesn’t it? Graduate students are (for the most part) smart and driven. So what sets you apart in a sea of ambitious smarties, all looking for similar jobs? Leveraging a pre-established connection to someone on the inside, that’s what.
The beauty of a conference is the people you meet and the network you build.
That’s a great advantage of grad school too; the ability to attend conferences and workshops that would be unavailable to non-students allows us to build those critical connections now.
Marketing Matters
 Scientists need to stop hiding behind jargon, and learn to communicate science to non-scientists in interesting ways. That’s the whole message of ComSciCon! By and large the people attending this conference already know that.
Scientists need to stop hiding behind jargon, and learn to communicate science to non-scientists in interesting ways. That’s the whole message of ComSciCon! By and large the people attending this conference already know that.
But herein lies a dilemma. As journalist/ ComSciCon panelist Phil McKenna said, (and as panelist Dietram Scheufele subsequently tweeted), “If you lose the jargon, you lose precision; if you don’t, you lose the audience.”
Is that a real tradeoff? Do we have to sacrifice information quality to make science accessible? I don’t think so. Science communicators shouldn’t sacrifice intellect for buzz. So, especially when challenging technical content is involved, the way we sell science matters tremendously.
If we step out of our labs and communicate, but we do it on blogs that no one reads, or on esoteric sites only known to other scientists, we’re just preaching to the choir. To reach people who don’t traditionally seek out science news, we need to find a means of communicating that retains challenging technical concepts, but shares those ideas in engaging, accessible ways with many, many people.
Unfortunately, ComSciCon didn’t host a panel on the specifics of effective marketing. We established that broadly accessible science communication is important, but didn’t set out actionable next steps.
It’s a pitfall of conferences in general I’d say, that panels based on the exchange of good ideas don’t necessarily translate into actionable steps back home. Because speakers are addressing large and diverse audiences, they tend to focus on somewhat generalized ideas, rather than laying out a protocol to change the field (or the world).
As a biologist, I like protocols. That said, even the most distinguished conference speaker can’t tell you what to do, or how to live your life. So how do we make positive change then? We write. We communicate. We just start, and en masse, find our voices and share them. Lose the jargon (but don’t assume the reader is an idiot) and communicate your research wherever you can.
It’s a process.
Be prepared to struggle. Be prepared to fail. The development of a career is an organic process and takes a looooong time.
Panelists overwhelmingly cited grit, rather than credentials, as key to their success. The tenacity to keep working and failing had carried many through years of career flux and financial insecurity, while they built up the network and experience to land more stable, permanent positions.
UGH, right? The prospect of protracted struggle and failure isn’t pretty, and certainly won’t increase the appeal of STEM outside of academia.
In terms of reach though, it seems that science communicators themselves are accessing broader audiences than ever before. As more and more trained researchers leave the academy, a new crop of science communication outlets has emerged. From quarterly publications like Nautilus to weekly podcasts like Radiolab, science communicators have more fingers in the pie than ever before.
Hopefully this means that if we continue to work together on smaller projects in the short term, there will be more large-scale opportunities in the long term.
Invest in the hours of your life, not in the idea of what you want to become.
Don’t get me wrong– it’s important to have long-term goals. Treat them as the cardinal directions on which you set the compass of your life. But then, rather than asking, “What do I want to end up doing?” ask, “What do I want to try next?” and “How can I put myself in a position to do that?”
If you invest in the moment and pursue the things you can’t help doing, you’re apt to take the inevitable challenges in stride, and to keep pushing through those early knee scrapes.
Not everyone has the privilege to do what he or she loves (financial security be damned), but if you are one of the lucky few with the opportunity to chase a not-so-lucrative passion, capitalize on that luck. Use it to build a more scientifically literate society. Use it to increase interest in science. Use it to build the foundations of a more diverse future in STEM!
–Amy McDermott